Microtubules Tool (3D)¶
The tool measures the total length of the microtubules in a 3D image. An example image can be found here.
License: CeCILL-C
Getting Started¶
To install the tools, drag the link MRI_Microtubules_Tools.ijm to the FIJI launcher window, save it under macros/toolsets in the FIJI installation and restart FIJI.
Select the "MRI_Microtubules_Tools" toolset from the >> button of the ImageJ launcher.

- the first button opens this help-page
- the m button measures the length of the microtubules in the active image
- the v button allows to measure the volume of a 3D object in a given channel of the image
Options¶
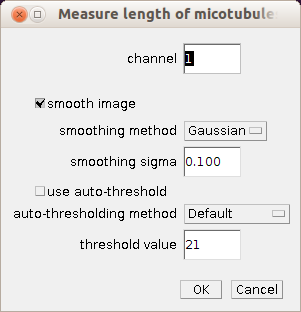
Right-click on the m-button to open the options dialog of the microtubules tool. Right-click on the v-button to open, the options dialog of the volume measurement tool.
 |
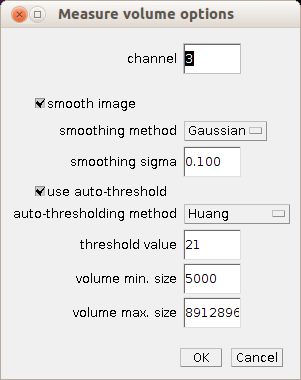 |
- channel: the number (1-) of the channel that contains the microtubules
- smooth image: if checked a smoothing filter is applied to the image
- smoothing method: the filter that is used for the smoothing if "smooth image" is checked
- smoothing sigma: the size of the smoothing filter if "smooth image" is checked and the method is either Gaussian or Uniform
- use auto-threshold: if checked auto-thresholding is used, otherwise a fixed threshold value is used
- auto-thresholding method: the auto-thresholding method that is used if "use auto-thresholding" is checked
- threshold value: the fixed threshold value that is used if "use auto-thresholding" is not checked
- volume min. size: the minimum volume of objects that are taken into account for the volume measurement
- volume max. size: the maximum volume of objects that are taken into account for the volume measurement
Method¶
The channel containing the microtubules is smoothed and and segmented by thresholding according to the selected options. The Skeletonize3D plugin written by Ignacio Arganda-Carreras is used to extract the centerlines of the microtubules. The AnalyzeSkeleton plugin from the same author provides the lengths of all the branches of the skeleton. The macro adds all branch lengths to calculate the total length of the microtubules.
Results¶
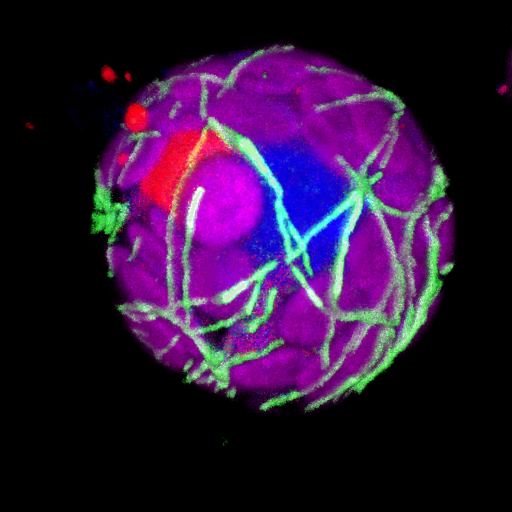 |
The input image contains the microtubules in the green channel which is the first channel in the hyperstack. |
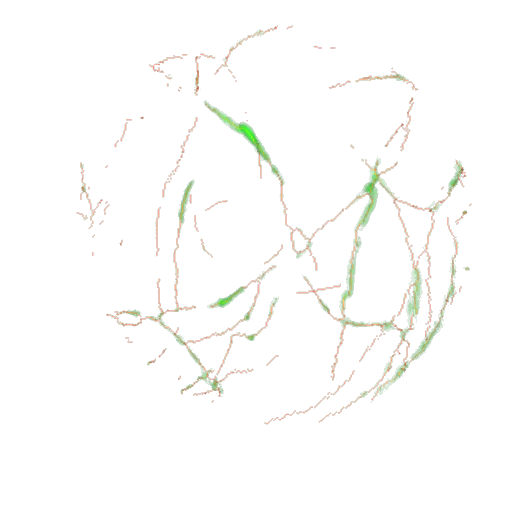 |
The skeleton (red) is saved into the subfolder skeletons of the input folder. It is shown as an overlay with the green channel in the ImageJ 3D viewer |
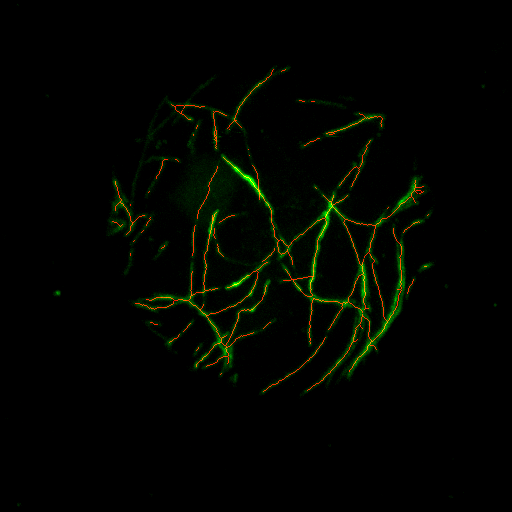 |
A projection of the overlay of the skeleton and the green channel is shown. The projection is saved into the subfolder skeletons-control of the input folder |
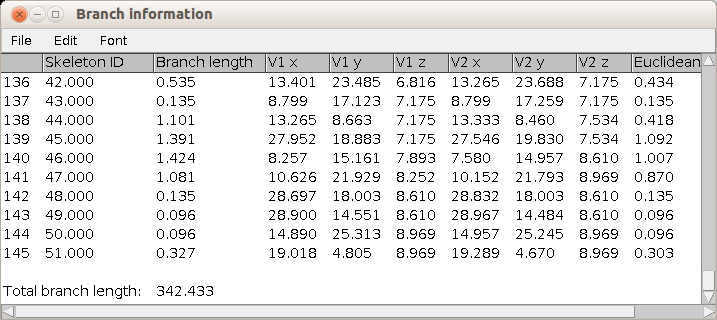 |
The total length of the skeleton is measured |
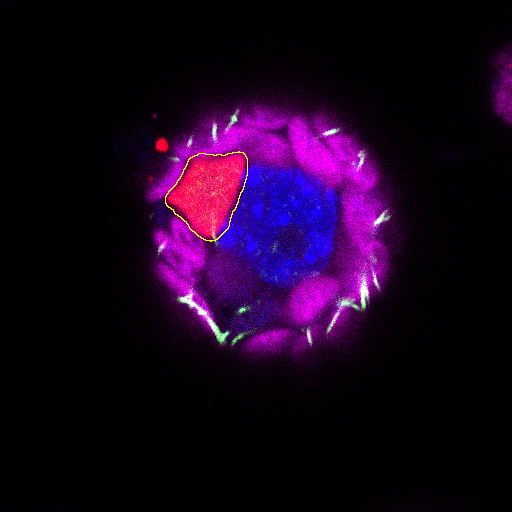 |
control image for the volume measurement - the measured area is drawn on each slice on which the object is present |
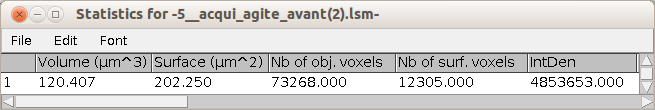 |
the results table of the volume measurement |